Google Scholar: Link
Orcid: Link
First author publications
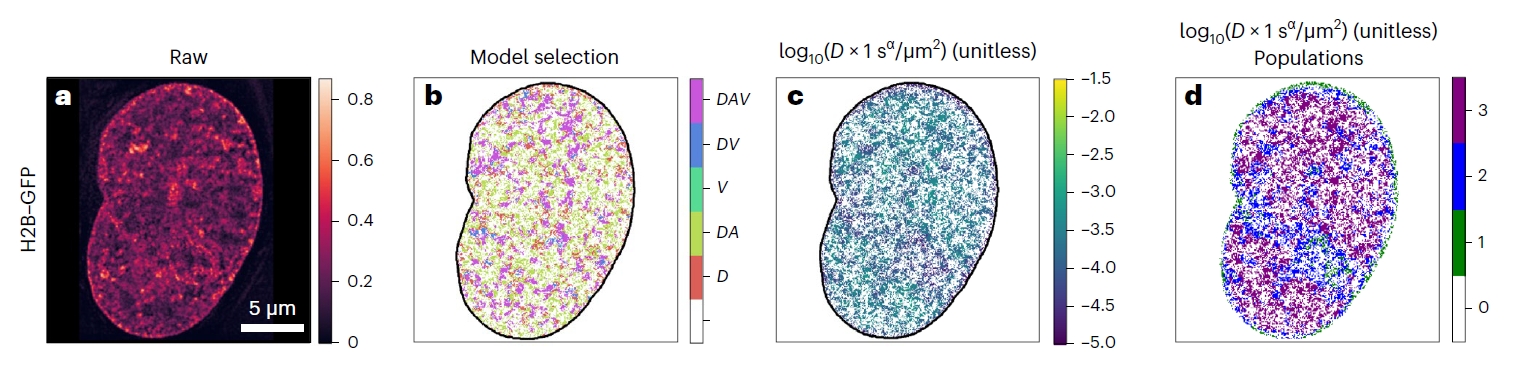
[1] Valades-Cruz C. A.*, Barth R.*, Abdellah M.*, Shaban H. A. Genome-wide analysis of the biophysical properties of chromatin and nuclear proteins in living cells with Hi-D. Nature Protocols (2025).
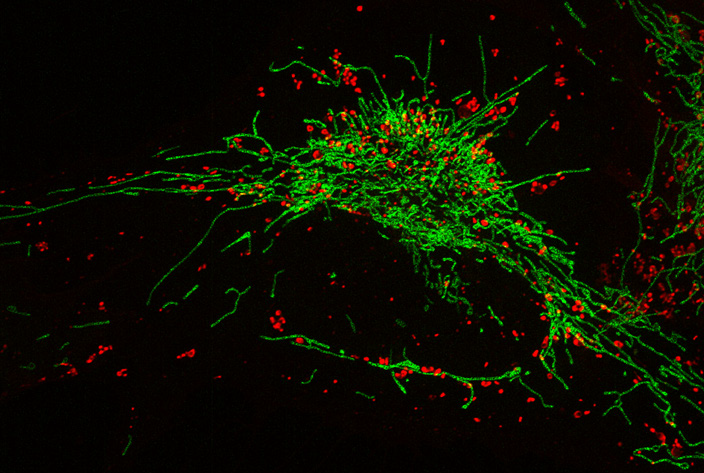
[2] Papereux S.*, Leconte L.*, Valades-Cruz C. A.*, Liu T., Dumont J., Chen Z., Salamero J., Kervrann C., Badoual A. DeepCristae, a CNN for the restoration of mitochondria cristae in live microscopy images. Communications Biology (2025)
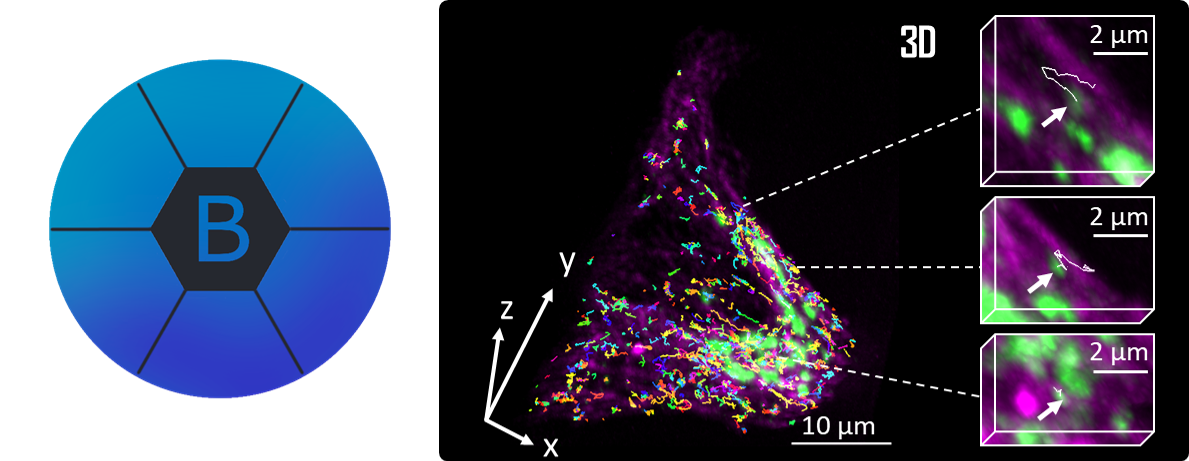
[3] Prigent S.*, Valades-Cruz C. A.*, Leconte L.*, Maury L., Salamero J., Kervrann C. BioImageIT: Open-source framework for integration of image data-management with analysis. Nature Methods (2022).
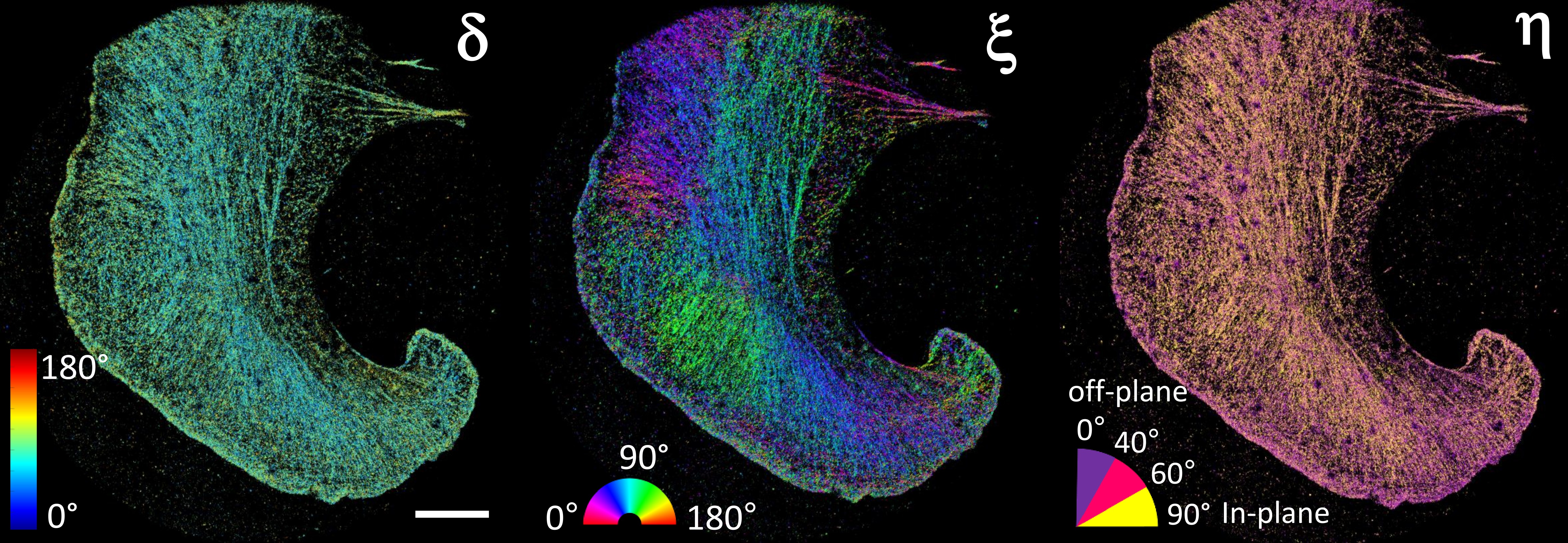
[4] Vaz Rimoli C.*, Valades-Cruz C. A.*, Curcio V., Mavrakis M., Brasselet S. 4polar-STORM polarized super-resolution imaging of actin filament organization in cells. Nature Communications (2022).
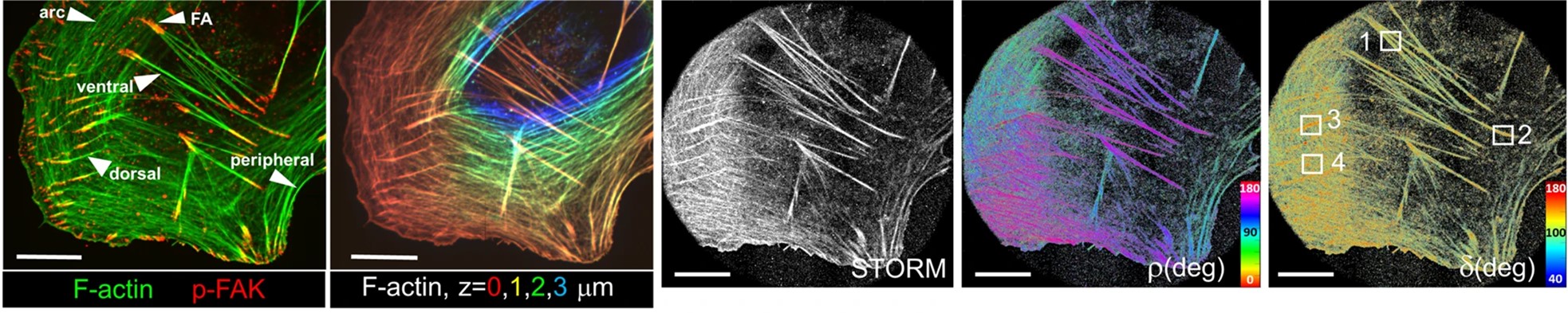
[5] Shaban H.*, Valades-Cruz C. A.* , Savatier J., Brasselet S. Polarized super-resolution structural imaging inside amyloid fibrils using Thioflavine T. Scientific Reports (2017).
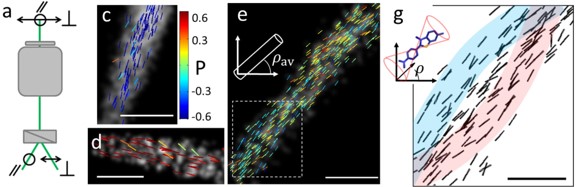
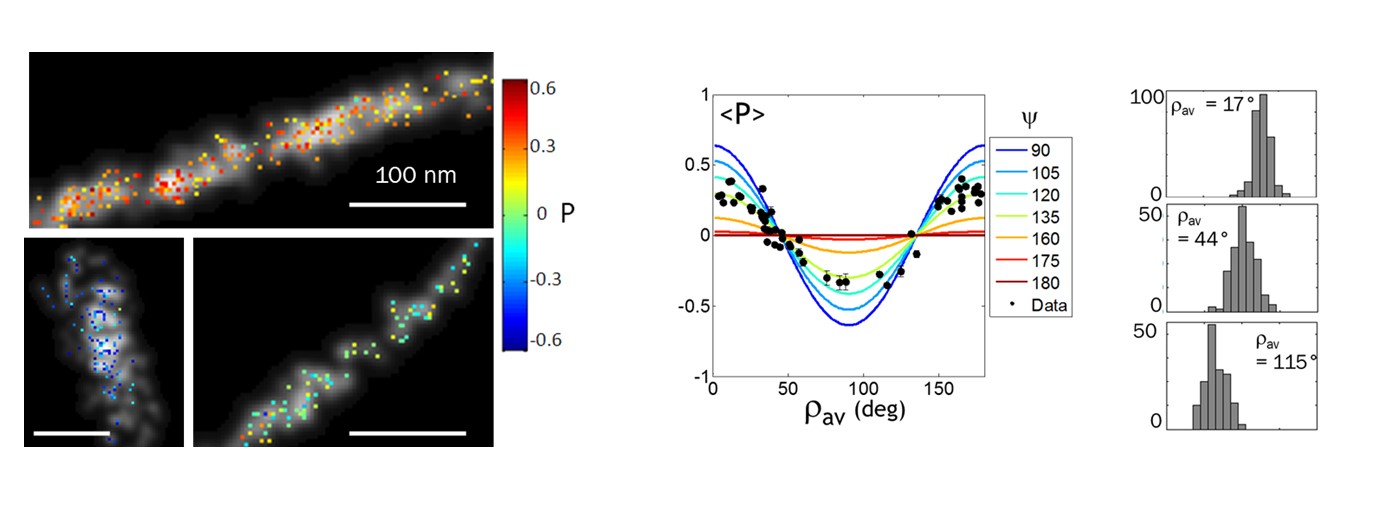
[6] Valades-Cruz C. A.*, Shaban H.* , Kress A., Bertaux N., Monneret S., Mavrakis M., Savatier J., Brasselet S. Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. PNAS (2016).
Corresponding author publications
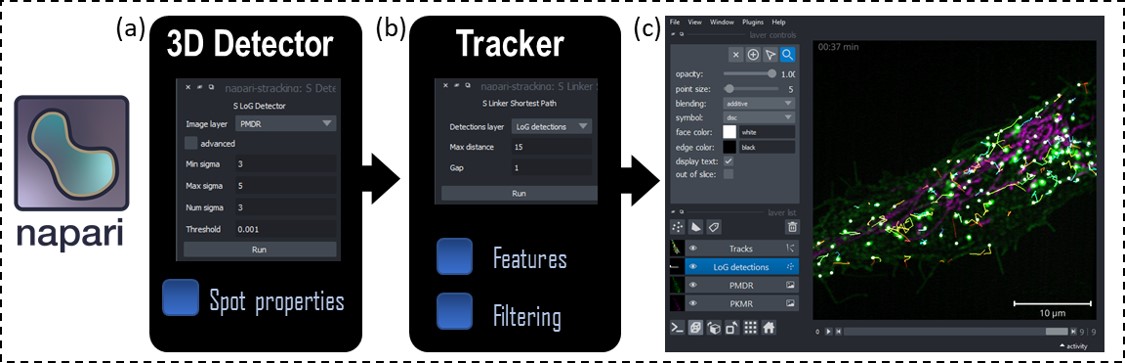
[1] Prigent S., Valades-Cruz C. A., Leconte L., Salamero J., Kervrann C. STracking: a free and open-source Python library for particle tracking and analysis. Bioinformatics (2022).
Other publications
[1] Shafaq-Zadah M., Dransart E., Hamitouche I., Wunder C., Chambon V., Valades-Cruz C. A., Leconte L., Sarangi N.K., Robinson J., Bai S-K., Regmi R., Di Cicco A., Hovasse A., Bartels R., Nilsson U. J., Cianférani-Sanglier S., Leffler H., Keyes T. E., Lévy D., Raunser S., Roderer D., Johannes L. Spatial N-glycan rearrangement on α5β1 integrin nucleates galectin-3 oligomers to determine endocytic fate. Nature Communications (2025)
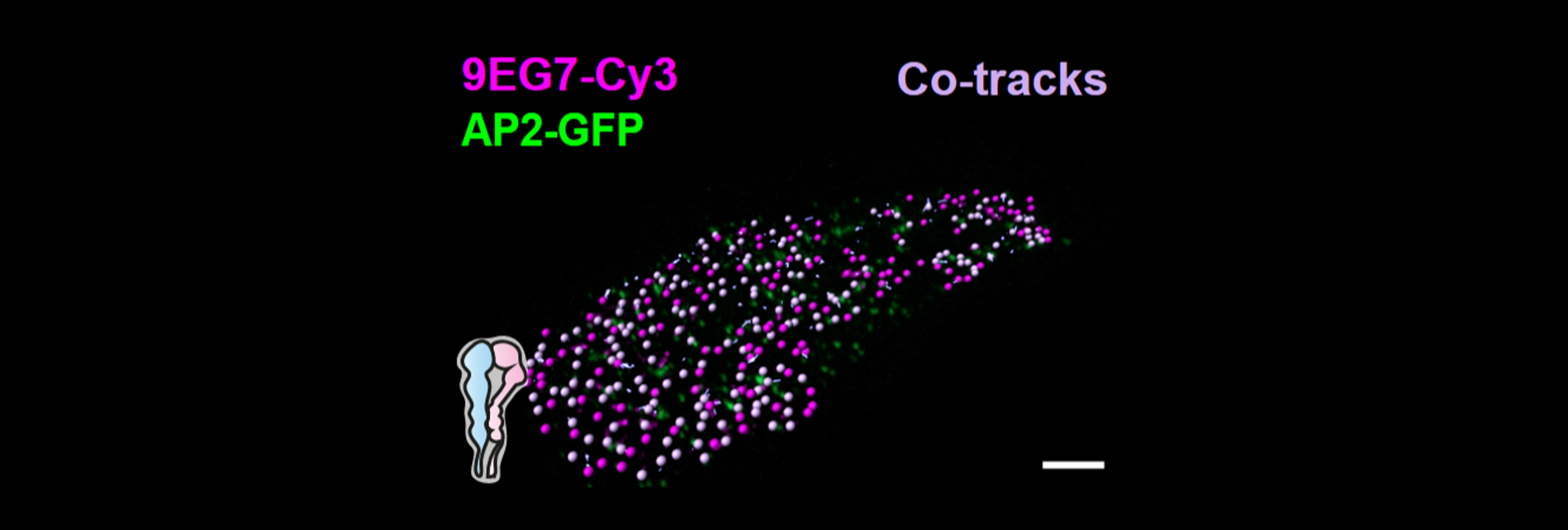
[2] MacDonald E., Forrester A., Valades-Cruz C. A., Madsen T. D., Hetmanski J. H. R., Dransart E., Ng Y., Godbole R., Shp A. A., Leconte L., Chambon V., Ghosh D., Pinet A., Bhatia D., Lombard B., Loew D., Larsen M. R., Leffler H., Lefeber D. J., Clausen H., Blangy A., Caswell P., Shafaq-Zadah M., Mayor S., Weigert R., Wunder C., Johannes L. Growth factor-triggered de-sialylation controls glycolipid-lectin-driven endocytosis. Nature Cell Biology (2025)
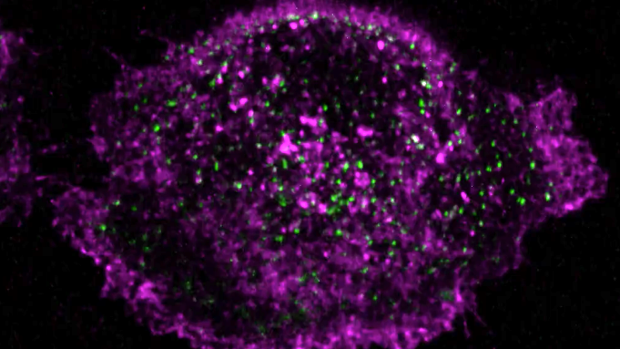
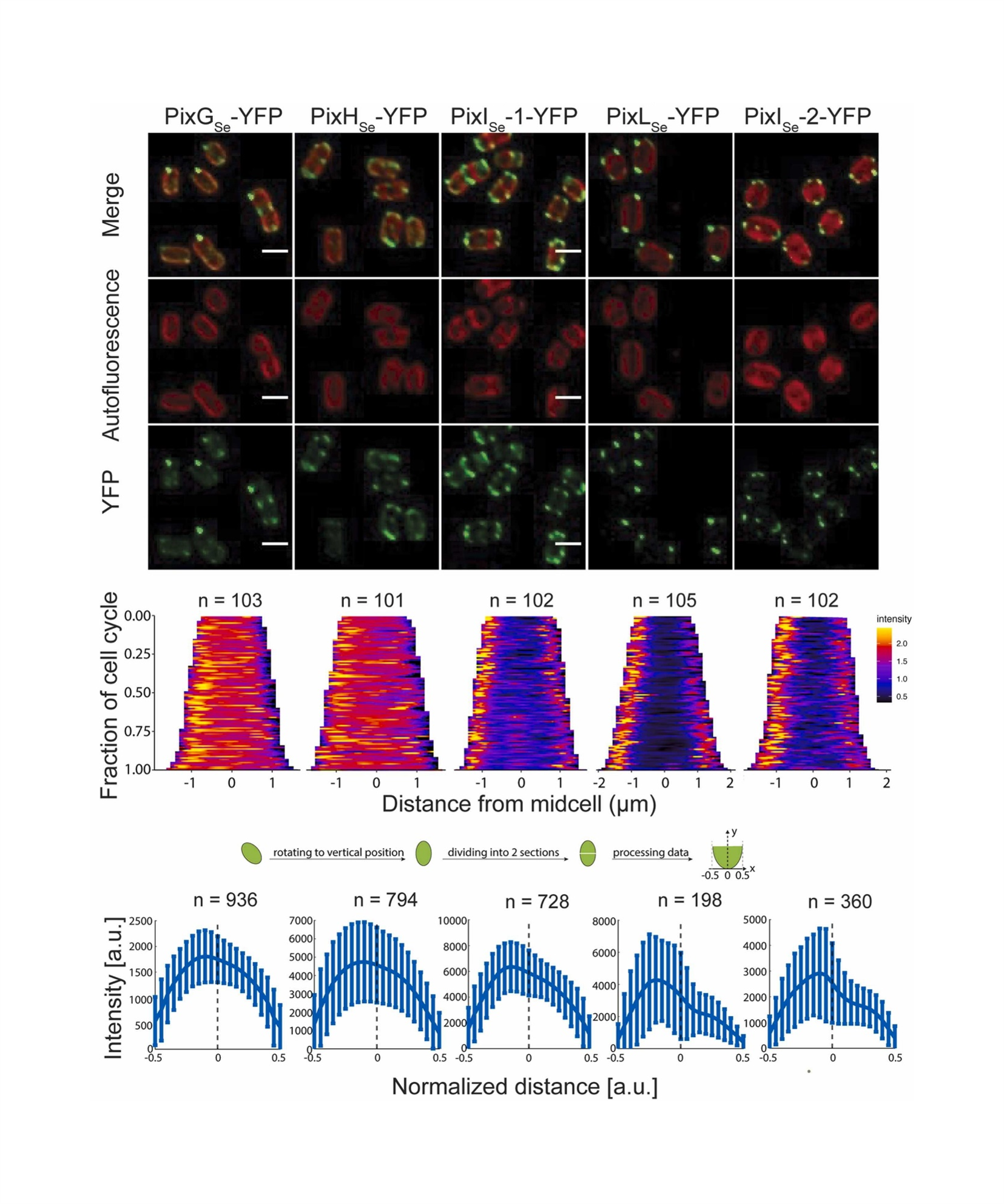
[3] Li S-Y., He C., Valades-Cruz C. A., Zhang C-C., Yang Y. Phototactic signaling network in rod-shaped cyanobacteria: a study on Synechococcus elongatus UTEX 3055. Microbiological Research (2025)
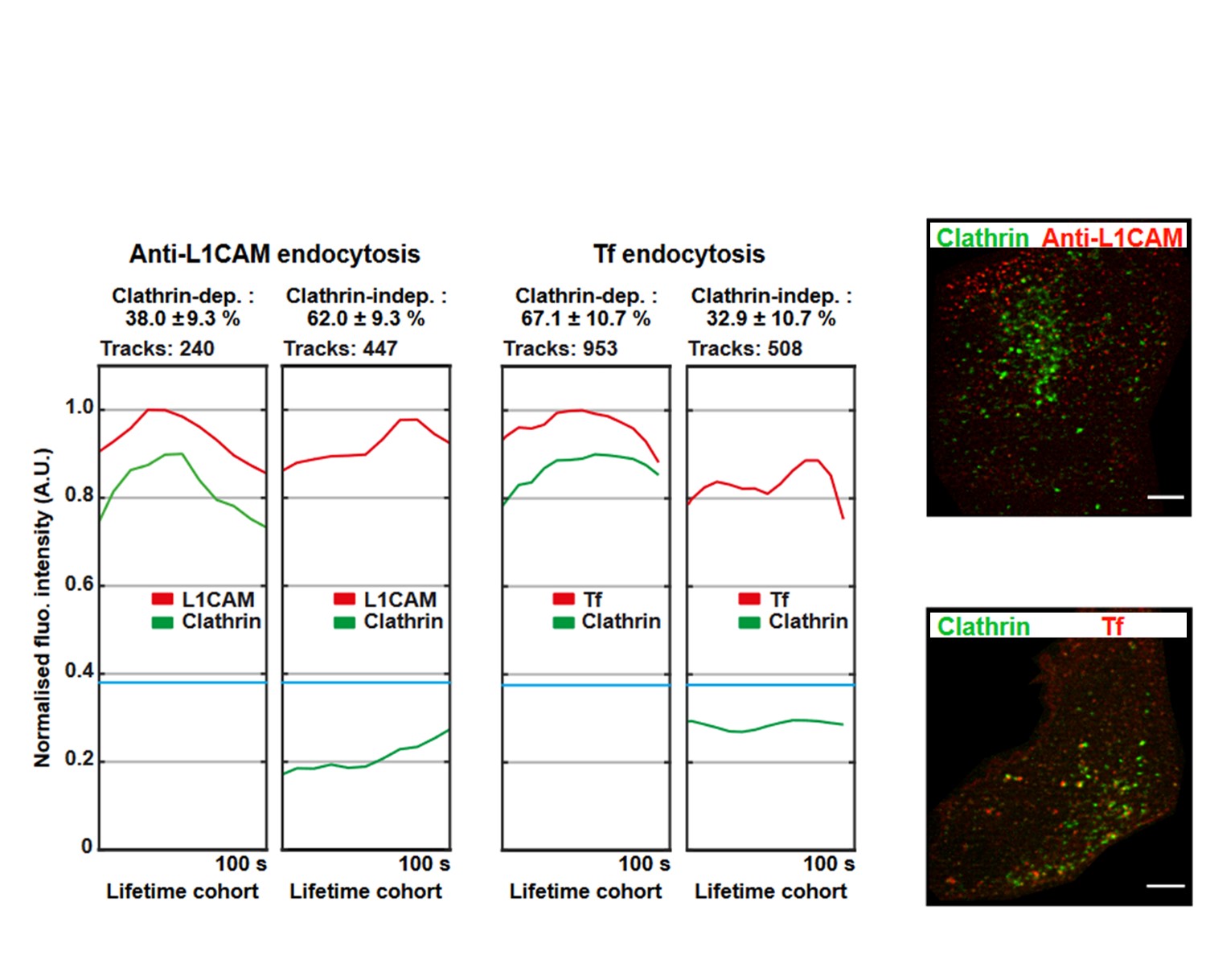
[4] Lemaigre C., Ceuppens A., Valades-Cruz C. A., Ledoux B., Vanbeneden B., Hassan M., Zetterberg F. R., Nilsson U. J., Johannes L., Wunder C., Renard H.-F. and Morsomme P. N-BAR and F-BAR proteins – Endophilin-A3 and PSTPIP1 – control clathrin-independent endocytosis of L1CAM. Traffic (2023)
[5] Prigent S., Nguyen H-N., Leconte L., Valades-Cruz C. A., Hajj B., Salamero J., Kervrann C. SPITFIR(e): a supermaneuverable algorithm for fast denoising and deconvolution of 3D fluorescence microscopy images and videos. Scientific Reports (2023)
[6] Forrester A., Rathjen S., Garcia-Castillo M. D., Bachert C., Couhert A., Tepshi L., Pichard S., Martinez J., Munier M., Sierocki R., Renard H. F., Valades-Cruz C. A., Dingli F., Loew D., Lamaze C., Cintrat J. C., Linstedt A., Gillet D., Barbier J., Johannes L. Functional Dissection of the Retrograde Shiga Toxin Trafficking Inhibitor Retro-2. Nature Chemical Biology (2020).
[7] Renard H-F., Tyckaert F., Lo Giudice C., Hirsch T., Valades-Cruz C. A., Lemaigre C., Shafaq-Zadah M., Wunder C., Wattiez R., Johannes L., van der Bruggen P., Alsteens D., Morsomme P. Endophilin-A3 and Galectin-8 control the clathrin-independent endocytosis of CD166. Nature Communications (2020).
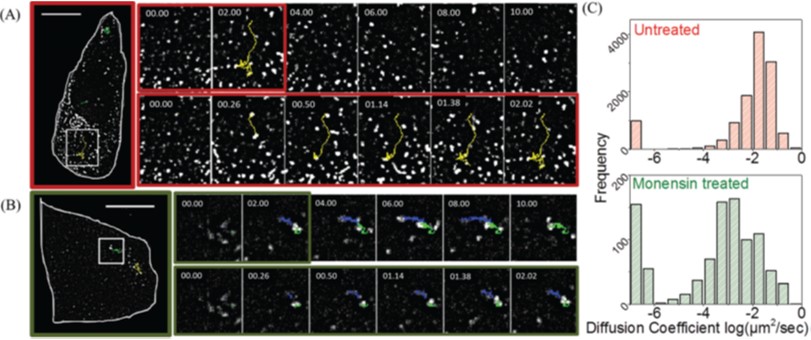
[8] Briane V., Vimond M., Valades-Cruz C. A., Salomon A., Wunder C., Kervrann C. A sequential algorithm to detect diffusion switching along intracellular particle trajectories. Bioinformatics (2020).
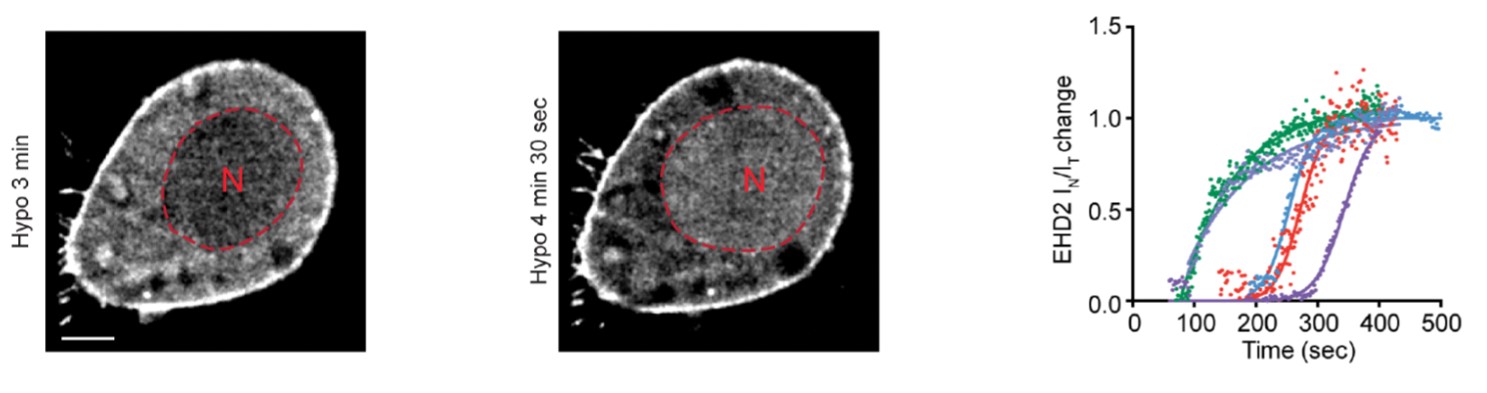
[9] Torrino S., Shen W., Blouin C., Kailasam Mani S., Viaris de Lesegno C., Bost P., Grassard A., Köster D., Valades-Cruz C. A., Chambon V., Johannes L., Pierobon P., Soumelis V., Coirault C., Vassilopoulos S., Lamaze C. EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. J Cell Biol. (2018).
[10] Banerjee A., Grazon C., Pons T., Bhatia D., Valades-Cruz C. A., Johannes L., Krishnan Y., Dubertret B. A Novel Type of Quantum Dot–Transferrin Conjugate Using DNA Hybridization Mimics Intracellular Recycling of Endogenous Transferrin. Nanoscale (2018).
Reviews, Perspective & Comments
[1] Valades-Cruz C. A.*, Leconte L.*, Fouche G.*, Blanc T.*, Van Hille N.*, Fournier K., Laurent T., Gallean B., Deslandes F., Hajj B., Faure E., Argelaguet F., Trubuil A., Isenberg T., Masson J-B., Salamero J., Kervrann C. Challenges of intracellular visualization using virtual and augmented reality. Frontiers in Bioinformatics (2022).

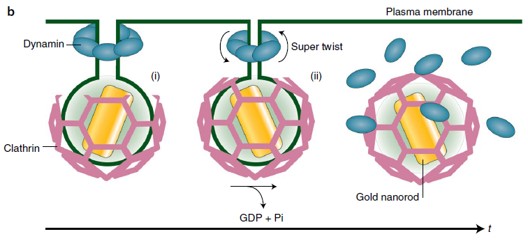
[2] Johannes L., Valades-Cruz C. A. The final twist in endocytic membrane scission. Nature Cell Biology (2021).
Proceedings
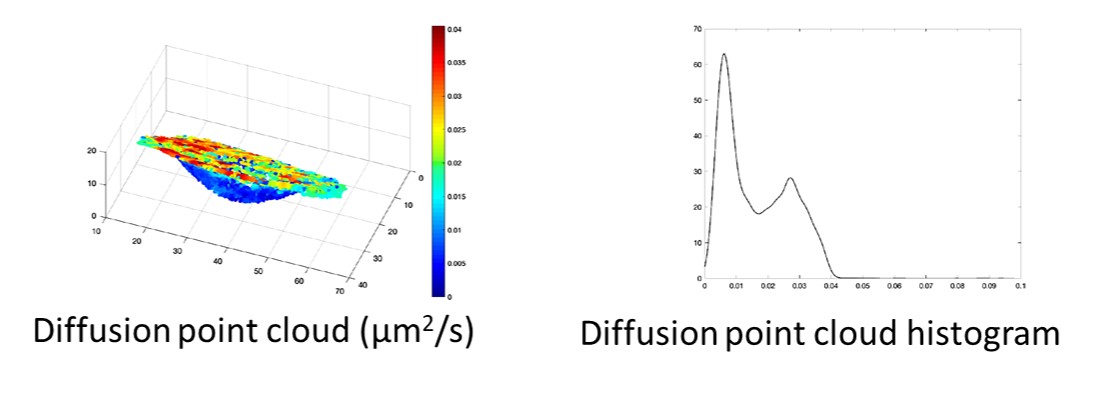
[1] Salomon A., Valades-Cruz C. A., Leconte L., Kervrann C. Dense Mapping of Intracellular Diffusion and Drift from Single-Particle Tracking Data Analysis, ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 2020, pp. 966-970.
Preprints and submitted papers
[1] Senthil Kumar C. S.*, Valades Cruz C. A.*, Sison M. *, Vesga A. G., Rey-Barroso J., Curcio V., Alemán-Castañeda L. A., Alonso M. A., Poincloux R., Mavrakis M., Brasselet S. 4polar3D: Single molecule 3D orientation imaging of dense actin networks using ratiometric polarization splitting. bioRxiv (2025)